Zhang Lichi's Group Publishes Seven Papers at MICCAI 2023
October 20, 2023
The International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) stands as a premier international academic conference that bridges the domains of Medical Image Computing (MIC) and Computer Assisted Intervention (CAI). At this year's MICCAI 2023, held in Vancouver, Canada, the team from the School of Biomedical Engineering at Shanghai Jiao Tong University, led by Professor Zhang Lichi's group, made a significant contribution by publishing a total of seven papers. Notably, three of these papers were accepted in advance, ranking within the top 14% of submissions. The research covered various domains, including brain imaging, musculoskeletal imaging, and pathological image analysis, showcasing the team's diverse and fruitful research outcomes.
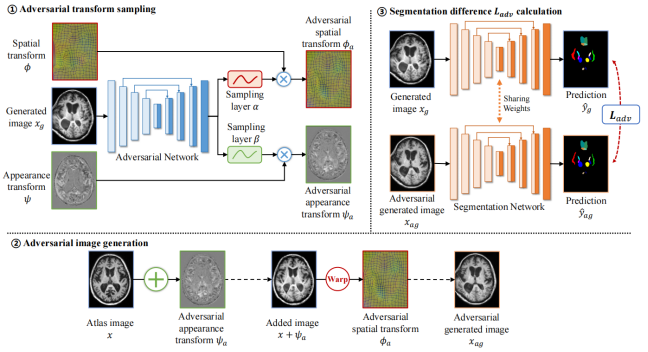
Brain segmentation of patients with severe traumatic brain injuries (sTBI) is essential for clinical treatment, but fully-supervised segmentation is limited by the lack of annotated data. One-shot segmentation based on learned transformations (OSSLT) has emerged as a powerful tool to overcome the limitations of insufficient training samples, which involves learning spatial and appearance transformations to perform data augmentation, and learning segmentation with augmented images. However, current practices face challenges in the limited diversity of augmented samples and the potential label error introduced by learned transformations. In this paper, we propose a novel one-shot traumatic brain segmentation method that surpasses these limitations by adversarial training and uncertainty rectification. The proposed method challenges the segmentation by adversarial disturbance of augmented samples to improve both the diversity of augmented data and the robustness of segmentation. Furthermore, potential label error introduced by learned transformations is rectified according to the uncertainty in segmentation. We validate the proposed method by the one-shot segmentation of consciousness-related brain regions in traumatic brain MR scans. Experimental results demonstrate that our proposed method has surpassed state-of-the-art alternatives. Code is available at https://github.com/hsiangyuzhao/TBIOneShot.
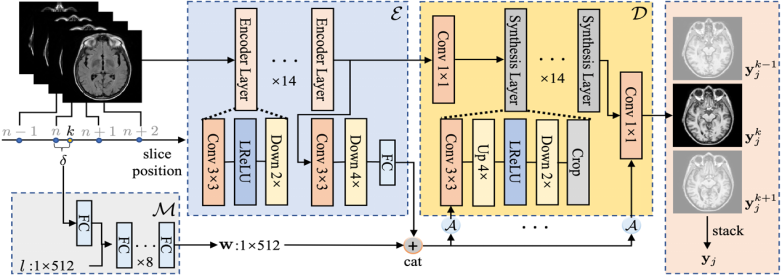
Cross-modality synthesis (CMS) and super-resolution (SR) have both been extensively studied with learning-based methods, which aim to synthesize desired modality images and reduce slice thickness for magnetic resonance imaging (MRI), respectively. It is also desirable to build a network for simultaneous cross-modality and super-resolution (CMSR) so as to further bridge the gap between clinical scenarios and research studies. However, these works are limited to specific fields. None of them can flexibly adapt to various combinations of resolution and modality, and perform CMS, SR, and CMSR with a single network. Moreover, alias frequencies are often treated carelessly in these works, leading to inferior detail-restoration ability. In this paper, we propose Alias-Free Co-Modulated network (AFCM) to accomplish all the tasks with a single network design. To this end, we propose to perform CMS and SR consistently with co-modulation, which also provides the flexibility to reduce slice thickness to various, non-integer values for SR. Furthermore, the network is redesigned to be alias-free under the Shannon-Nyquist signal processing framework, ensuring efficient suppression of alias frequencies. Experiments on three datasets demonstrate that AFCM outperforms the alternatives in CMS, SR, and CMSR of MR images. Our codes are available at https://github.com/zhiyuns/AFCM.
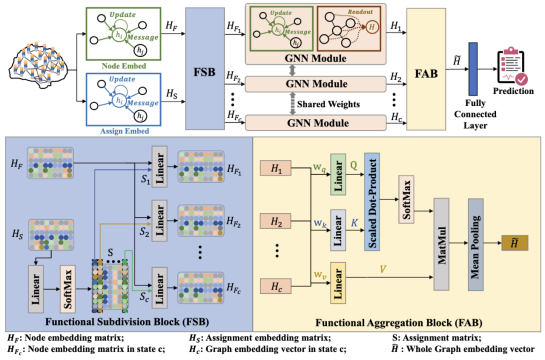
Different functional configurations of the brain, also named as "brain states", reflect a continuous stream of brain cognitive activities. These distinct brain states can confer heterogeneous functions to brain networks. Recent studies have revealed that extracting information from functional brain networks is beneficial for neuroscience analysis and brain disorder diagnosis. Graph neural networks (GNNs) have been demonstrated to be superior in learning network representations. However, these GNN-based methods have few concerns about the heterogeneity of brain networks, especially the heterogeneous information of brain network functions induced by intrinsic brain states. To address this issue, we propose a learnable subdivision graph neural network (LSGNN) for brain network analysis. The core idea of LSGNN is to implement a learnable subdivision method to encode brain networks into multiple latent feature subspaces corresponding to functional configurations, and realize the feature extraction of brain networks in each subspace, respectively. Furthermore, considering the complex interactions among brain states, we also employ the self-attention mechanism to acquire a comprehensive brain network representation in a joint latent space. We conduct experiments on a publicly available dataset of cognitive disorders. The results affirm that our approach can achieve outstanding performance and also instill the interpretability of the brain network functions in the latent space. Our code is available at https://github.com/haijunkenan/LSGNN.
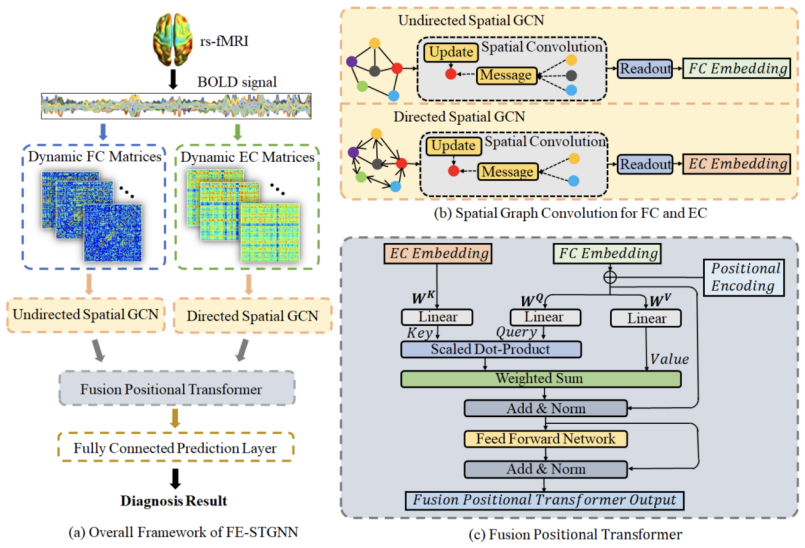
Brain connectivity patterns such as functional connectivity (FC) and effective connectivity (EC), describing complex spatio-temporal dynamic interactions in the brain network, are highly desirable for mild cognitive impairment (MCI) diagnosis. Major FC methods are based on statistical dependence, usually evaluated in terms of correlations, while EC generally focuses on directional causal influences between brain regions. Therefore, comprehensive integration of FC and EC with complementary information can further extract essential biomarkers for characterizing brain abnormality. This paper proposes Spatio-Temporal Graph Neural Network with Dynamic Functional and Effective Connectivity Fusion (FE-STGNN) for MCI diagnosis using resting-state fMRI (rs-fMRI). First, dynamic FC and EC networks are constructed to encode the functional brain networks into multiple graphs. Then, spatial graph convolution is employed to process spatial structural features and temporal dynamic characteristics. Finally, we design the position encoding-based cross-attention mechanism, which utilizes the causal linkage of EC during time evolution to guide the fusion of FC networks for MCI classification. Qualitative and quantitative experimental results demonstrate the significance of the proposed FE-STGNN method and the benefit of fusing FC and EC, which achieves
of MCI classification accuracy and outperforms state-of-the-art methods. Our code is available at https://github.com/haijunkenan/FE-STGNN.
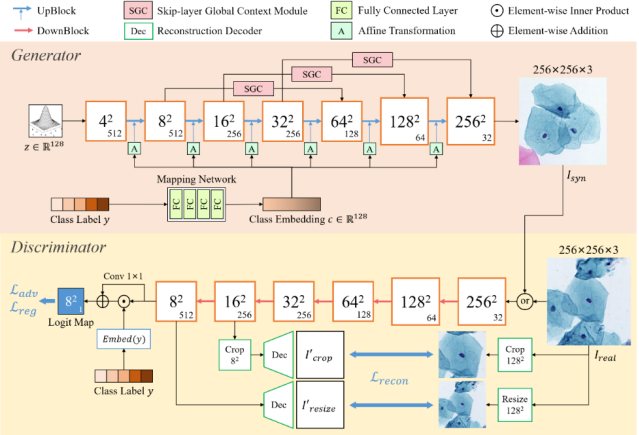
Automatic examination of thin-prep cytologic test (TCT) slides can assist pathologists in finding cervical abnormality for accurate and efficient cancer screening. Current solutions mostly need to localize suspicious cells and classify abnormality based on local patches, concerning the fact that whole slide images of TCT are extremely large. It thus requires many annotations of normal and abnormal cervical cells, to supervise the training of the patch-level classifier for promising performance. In this paper, we propose CellGAN to synthesize cytopathological images of various cervical cell types for augmenting patch-level cell classification. Built upon a lightweight backbone, CellGAN is equipped with a non-linear class mapping network to effectively incorporate cell type information into image generation. We also propose the Skip-layer Global Context module to model the complex spatial relationship of the cells, and attain high fidelity of the synthesized images through adversarial learning. Our experiments demonstrate that CellGAN can produce visually plausible TCT cytopathological images for different cell types. We also validate the effectiveness of using CellGAN to greatly augment patch-level cell classification performance. Our code and model checkpoint are available at https://github.com/ZhenrongShen/CellGAN.
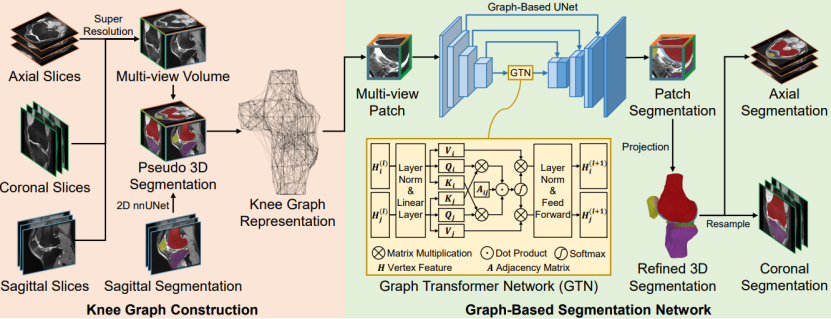
Magnetic Resonance Imaging (MRI) has become an essential tool for clinical knee examinations. In clinical practice, knee scans are acquired from multiple views with stacked 2D slices, ensuring diagnosis accuracy while saving scanning time. However, obtaining fine 3D knee segmentation from multi-view 2D scans is challenging, which is yet necessary for morphological analysis. Moreover, radiologists need to annotate the knee segmentation in multiple 2D scans for medical studies, bringing additional labor. In this paper, we propose the Cross-view Aligned Segmentation Network (CAS-Net) to produce 3D knee segmentation from multi-view 2D MRI scans and annotations of sagittal views only. Specifically, a knee graph representation is firstly built in a 3D isotropic space after the super-resolution of multi-view 2D scans. Then, we utilize a graph-based network to segment individual multi-view patches along the knee surface, and piece together these patch segmentations into a complete knee segmentation with help of the knee graph. Experiments conducted on the Osteoarthritis Initiative (OAI) dataset demonstrate the validity of the CAS-Net to generate accurate 3D segmentation.
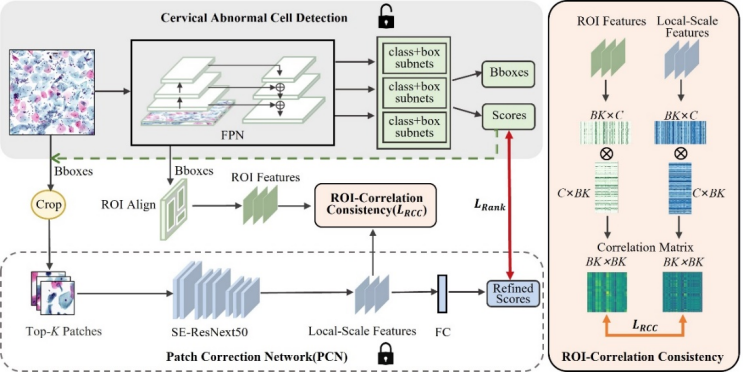
Automated detection of cervical abnormal cells from Thin-prep cytologic test (TCT) images is essential for efficient cervical abnormal screening by computer-aided diagnosis system. However, the detection performance is influenced by noise samples in the training dataset, mainly due to the subjective differences among cytologists in annotating the training samples. Besides, existing detection methods often neglect visual feature correlation information between cells, which can also be utilized to aid the detection model. In this paper, we propose a cervical abnormal cell detection method optimized by a novel distillation strategy based on local-scale consistency refinement. Firstly, we use a vanilla RetinaNet to detect top-K suspicious cells and extract region-of-interest (ROI) features. Then, a pre-trained Patch Correction Network (PCN) is leveraged to obtain local-scale features and conduct further refinement for these suspicious cell patches. We design a classification ranking loss to utilize refined scores for reducing the effects of the noisy label. Furthermore, the proposed ROI-correlation consistency loss is computed between extracted ROI features and local-scale features to exploit correlation information and optimize RetinaNet. Our experiments demonstrate that our distillation method can greatly optimize the performance of cervical abnormal cell detection without changing the detector’s network structure in the inference. The code is publicly available at https://github.com/feimanman/Cervical-Abnormal-Cell-Detection.