Hongjiang Wei Group: Deep Learning for Quantitative Susceptibility Mapping
January 01, 2023
1. Learning-based single-step quantitative susceptibility mapping reconstruction without brain extraction
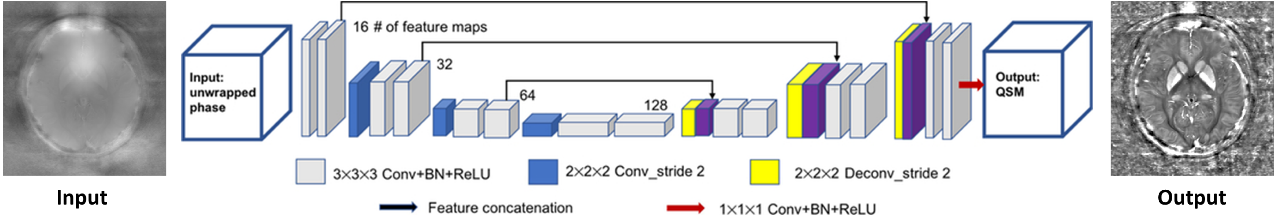
Quantitative susceptibility mapping (QSM) estimates the underlying tissue magnetic susceptibility from MRI gradient-echo phase signal and typically requires several processing steps. These steps involve phase unwrapping, brain volume extraction, background phase removal and solving an ill-posed inverse problem relating the tissue phase to the underlying susceptibility distribution. The resulting susceptibility map is known to suffer from inaccuracy near the edges of the brain tissues, in part due to imperfect brain extraction, edge erosion of the brain tissue and the lack of phase measurement outside the brain. This inaccuracy has thus hindered the application of QSM for measuring susceptibility of tissues near the brain edges, e.g., quantifying cortical layers and generating superficial venography. To address these challenges, we propose a learning-based QSM reconstruction method that directly estimates the magnetic susceptibility from total phase images without the need for brain extraction and background phase removal, referred to as autoQSM. The neural network has a modified U-net structure and is trained using QSM maps computed by a two-step QSM method. 209 healthy subjects with ages ranging from 11 to 82 years were employed for patch-wise network training. The network was validated on data dissimilar to the training data, e.g., in vivo mouse brain data and brains with lesions, which suggests that the network generalized and learned the underlying mathematical relationship between magnetic field perturbation and magnetic susceptibility. Quantitative and qualitative comparisons were performed between autoQSM and other two-step QSM methods. AutoQSM was able to recover magnetic susceptibility of anatomical structures near the edges of the brain including the veins covering the cortical surface, spinal cord and nerve tracts near the mouse brain boundaries. The advantages of high-quality maps, no need for brain volume extraction, and high reconstruction speed demonstrate autoQSM's potential for future application.
Link: https://pubmed.ncbi.nlm.nih.gov/31377323/
2. MoDL-QSM: Model-based deep learning for quantitative susceptibility mapping
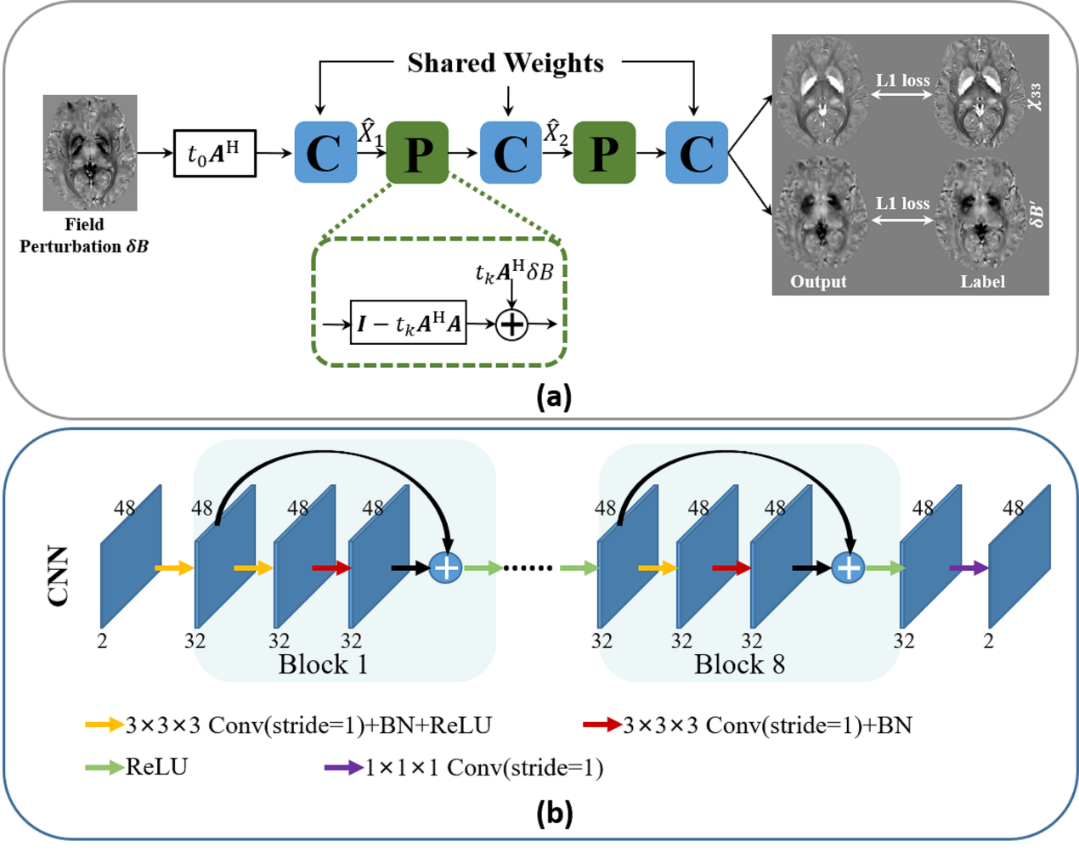
Quantitative susceptibility mapping (QSM) has demonstrated great potential in quantifying tissue susceptibility in various brain diseases. However, the intrinsic ill-posed inverse problem relating the tissue phase to the underlying susceptibility distribution affects the accuracy for quantifying tissue susceptibility. Recently, deep learning has shown promising results to improve accuracy by reducing the streaking artifacts. However, there exists a mismatch between the observed phase and the theoretical forward phase estimated by the susceptibility label. In this study, we proposed a model-based deep learning architecture that followed the STI (susceptibility tensor imaging) physical model, referred to as MoDL-QSM. Specifically, MoDL-QSM accounts for the relationship between STI-derived phase contrast induced by the susceptibility tensor terms (χ13, χ23 and χ33) and the acquired single-orientation phase. The convolutional neural networks are embedded into the physical model to learn a regularization term containing prior information. χ33 and phase induced by χ13 and χ23 terms were used as the labels for network training. Quantitative evaluation metrics were compared with recently developed deep learning QSM methods. The results showed that MoDL-QSM achieved superior performance, demonstrating its potential for future applications.
Link: https://pubmed.ncbi.nlm.nih.gov/34246768/
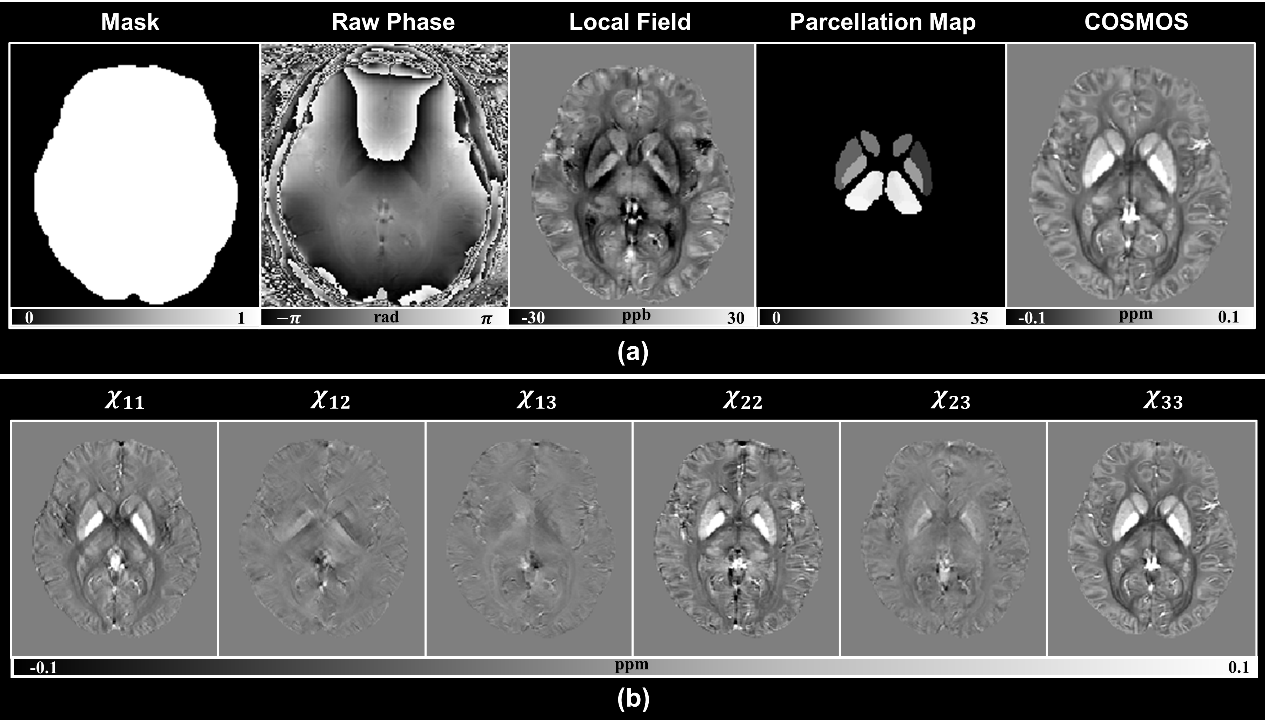
3. Towards in vivo ground truth susceptibility for single-orientation deep learning QSM: A multi-orientation gradient-echo MRI dataset
Recently, deep neural networks have shown great potential for solving dipole inversion of quantitative susceptibility mapping (QSM) with improved results. However, these studies utilized their limited dataset for network training and inference, which may lead to untrustworthy conclusions. Thus, a common dataset is needed for a fair comparison between different QSM reconstruction networks. Additionally, finding an in vivo reference susceptibility map that matches acquired single-orientation phase data remains an open problem. Susceptibility tensor imaging (STI) χ33 and Calculation of Susceptibility through Multiple Orientation Sampling (COSMOS) are considered reference susceptibility candidates. However, a large number of multi-orientation GRE data for both STI and COSMOS reconstruction are now unavailable for training supervised neural networks for QSM. In this study, we reported the largest multi-orientation dataset, to the best of our knowledge in the QSM research field, with a total of 144 scans from 8 healthy subjects collected using a 3D GRE sequence from the same MR scanner. In addition, the parcellation of deep gray matter is also provided for automatically extracting susceptibility values. Five recently developed deep neural networks, i.e., xQSM, QSMnet, autoQSM, LPCNN, and MoDL-QSM were performed on this dataset. This potential data source could provide a common framework and labels to test the accuracy and robustness of deep neural networks for QSM reconstruction. This dataset has the potential to provide a benchmark of reference susceptibility for the deep learning-based QSM methods. Additionally, the trained COSMOS-labeled and χ33-labeled networks were tested on the pathological data to explore their potential applications. The data together with deep gray matter parcellation maps are now publicly available via an open repository at https://osf.io/yfms7/, and the raw multi-orientation GRE data are also available at https://osf.io/y6rc3/.