


NatGenet |Li Li's team from the school of Biomedical Engineering reveals the mechanism and functions of oocyte epigenome
May 01, 2019
Li Li's research group, school of Biomedical Engineering, Shanghai Jiao Tong University, Jie Wei's research group, School of Life Science, Tsinghua University, and Li Wei's research group, Institute of Zoology, Chinese Academy of Sciences, In the journal Nature Genetics, published a long article titled "Setd2 Regulates Maternal Epigenome, Genomic Imprinting and Early Embryonic Development", on April 29, 2019.
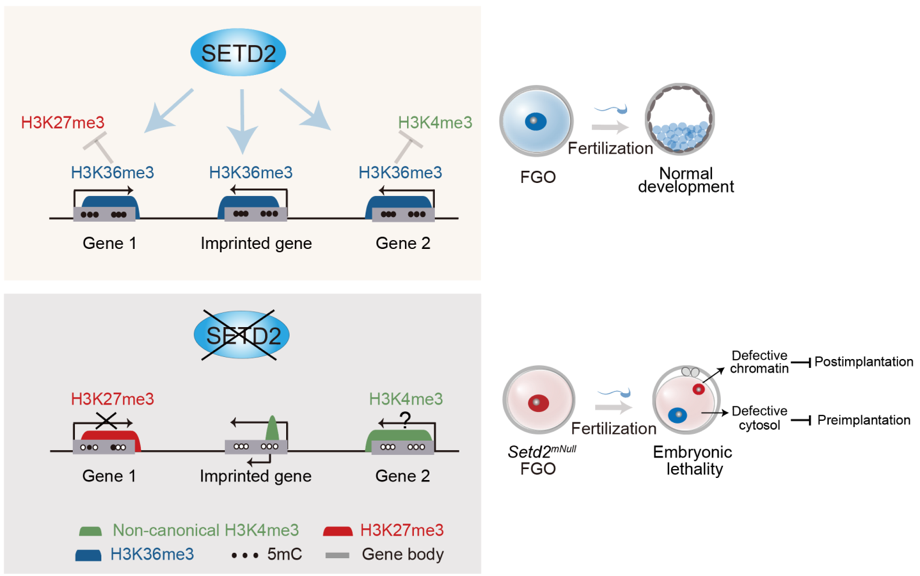
In this article, they first report that histone-lysine N-methyltransferase SETD2, an H3K36me3 methyltransferase, is a crucial regulator of the mouse oocyte epigenome. Deficiency in Setd2 leads to extensive alterations of the oocyte epigenome, including the loss of H3K36me3, failure in establishing the correct DNA methylome, invasion of H3K4me3 and H3K27me3 into former H3K36me3 territories and aberrant acquisition of H3K4me3 at imprinting control regions instead of DNA methylation. Importantly, maternal depletion of SETD2 results in oocyte maturation defects and subsequent one-cell arrest after fertilization. The preimplantation arrest is mainly due to a maternal cytosolic defect, since it can be largely rescued by normal oocyte cytosol. However, chromatin defects, including aberrant imprinting, persist in these embryos, leading to embryonic lethality after implantation. Thus, these data identify SETD2 as a crucial player in establishing the maternal epigenome that in turn controls embryonic development.
Epigenetic marks play critical roles in regulating chromatin state and gene expression in concert with DNA sequences. These modifications can be maintained through cell cycles to transmit the memory of cell identities. Abnormalities in the epigenome cause severe developmental defects and human diseases. Fertilization provides an opportunity for transmitting not only genetic information but also epigenetic information from parents to early embryos. Oocytes, in particular, exert a profound impact on embryogenesis by transmitting not only a large amount of RNAs and proteins but also certain epigenetic marks. For example, despite extensive epigenetic reprogramming after fertilization, oocyte-established DNA methylation at imprinted loci is stably inherited in embryos and enables allele-specific gene expression. Maternal depletion of DNA methyltransferases leads to postimplantation lethality. Similarly, emerging evidence indicates that maternally inherited histone modifications can also carry out crucial functions. For instance, oocyte-transmitted H3K27me3 can regulate allele-specific gene expression and X chromosome inactivation in early mouse embryos. These data demonstrate that the maternal epigenome plays vital roles in oogenesis and early embryogenesis.
Professor Wei Xie, Associate Researcher Li Li, and Professor Wei Li are the corresponding authors of this article. Dr. Xu Qianhua, PhD students Yunlong Xiang, Qiujun Wang and Dr. Leyun Wang are the co-first authors of this article. Cooperative laboratories include the Matthew C. Lorincz research group of the University of British Columbia, the Louis Lefebvre research group and the Wu Min research group of Wuhan University.
This research has been supported by the National Key R&D Program of China, the National Basic Research Program of China, the State Key Laboratory of Oncogenes and Related Genes, the Shanghai Science and Technology Commission, and an Innovation Research Plan from the Shanghai Municipal.