


Xianting Ding, Guan Ning Lin’s team proposed a fundamental analysis framework for single cell mass spectrometry data clustering method
December 30, 2019
On December 23, 2019, Professor Xianting Ding and Professor Lin Guanning, Shanghai Jiaotong University, jointly published teh article "A Comparison Framework and Guideline of Clustering Methods for Mass Cytometry Data" on Genome Biology. They clarifies the pros and cons of different single-cell mass spectrometry flow cytometry (CyTOF) cell population analysis methods and their application scenarios by three aspects of accuracy, coherence, and stability. It is the first time that an international top magazine reported Chinese mainland scholars’ work on the fields of data standardization and analysis methodology of single-cell mass spectrometry flow cytometry.
During the last decade, single-cell technology has progressed tremendously. With the ability to simultaneously measure multiple features at the single-cell level, biologists are now capable of depicting biological and pathological processes with unprecedented complexity. Mass cytometry, which is achieved with Cytometry by Time-OfFlight (CyTOF), is an advanced experimental technology that measures levels of multiple proteins (up to 40) in a large amount (usually several millions) of cells. The supreme ability to access a large panel of proteins simultaneously makes CyTOF useful in drug optimization, vaccine development, and disease marker discovery. Compared to the well-known technology of single-cell
RNA-sequencing (scRNA-seq), which processes on average tens of thousands to hundreds of thousands of cells, CyTOF achieves a higher throughput (on average up to millions of cells) and classifies cells from a mixture into distinct subtypes based on expression levels of their surface antigen. Cells are first stained by antibodies labeled with metal isotopes and then travel through a time-of-flight mass spectrometer, where the density of each isotope label is quantified.
In this paper, they compared the performance of nine popular clustering methods in three categories—precision, coherence, and stability—using six independent datasets. This comparison would allow cytometry scientists to choose the most appropriate tool with clear answers to the following questions: (1) How does one select between unsupervised and semi-supervised tools? (2) How does one choose the most suitable unsupervised or semi-supervised tool in its category?
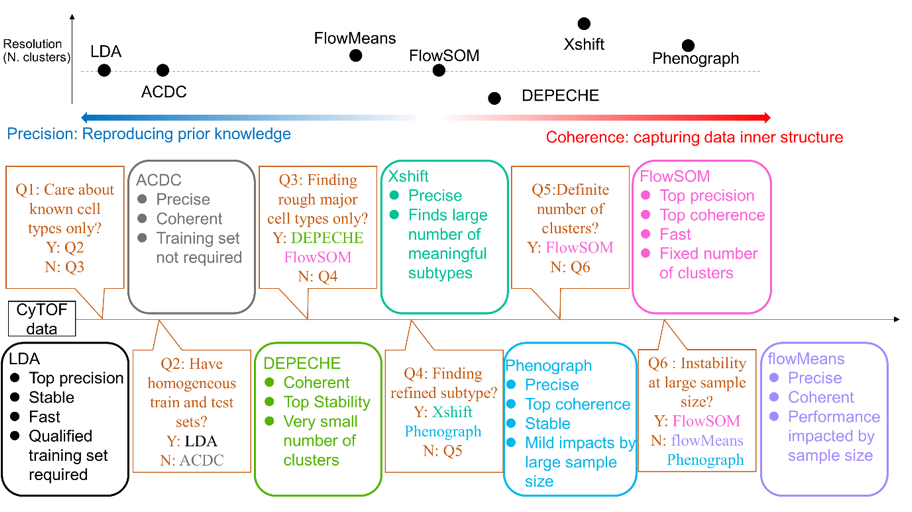
Fig.1 Summary of characteristics of each tool and suggested decision pipeline to choose the right tool
This paper provides a selection decision tree for cell clustering methods for researchers in the field of single-cell mass spectrometry, especially novices and those without a bioinformatics background.
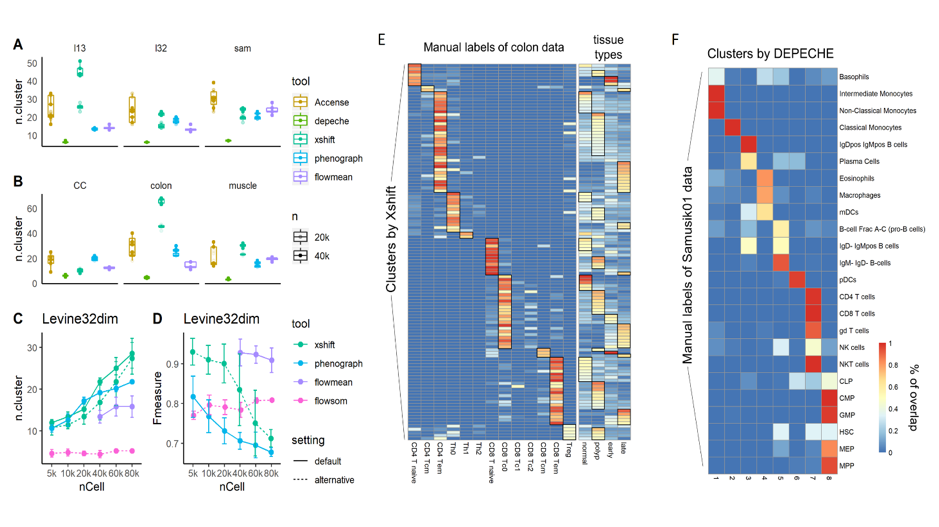
Fig.2 Stability analysis of clustering method
Dr. Xiao Liu and Dr. Weichen Song are the first authors of the paper. Professor Xianting Ding and Professor Guanning Lin are the corresponding authors. This research is supported by the Shanghai Municipal Science and Technology (2017SHZDZX01), National Natural Science Foundation of China (81871448,81671328), National Key Research and Development Program of China(2017ZX10203205-006-002), Innovation Research Plan supported by Shanghai Municipal Education Commission (ZXWF082101), National Key R&D Program of China (2017YFC0909200).
Article Link: https://doi.org/10.1186/s13059-019-1917-7